Introduction
The 2023 Nobel Prize in Physiology or Medicine was awarded jointly to Katalin Karikó and Drew Weissman1 ‘for their discoveries concerning nucleoside base modifications that enabled the development of effective mRNA vaccines against COVID-19.’ It is their technology which underpins both of the Pfizer-BioNTech and Moderna COVID-19 vaccines that so many of us have taken doses of over the past 3 years. You may, quite rightly, be wondering why an article about the 2023 Nobel Prize in Physiology or Medicine is appearing in the journal of the New Zealand Institute of Chemistry. Partly this is due to my previous two Chemistry in New Zealand articles on the 20212 and 20223 chemistry Nobel prizes, which suggested that Karikó and Weissman may have been chemistry awardees for either of those years. It is also in part because at the end of the day the brilliance of the mRNA vaccine development for which the 2023 Physiology/Medicine Nobel was awarded relies on chemistry at its heart, and in particular some beautifully nuanced sugar chemistry, which fools the immune system. Finally this is also a beautiful and inspirational story of ‘rags to riches’ in which determination and fundamental academic interest overcame funding bias and disparaging criticism from anonymous peer reviewers that should be told as widely as possible.
Of course, this prize is also extremely topical. The COVID-19 pandemic has had numerous unprecedented effects on society. One of these was thankfully to raise public awareness and appreciation of science. Never before has the person on the street been so acutely aware of vaccine development and implementation. It has also resulted in the self-identification of a considerable number of people within society as anti-vaxxers, conspiracy theorists and science deniers. In this latter respect it appears that global educational systems still have a considerable amount of work to do. However, let us forget about those Luddites, and discuss this year’s awardees, but before we do so we should remind ourselves of some basic molecular biology and immunology, as these are central to the 2023 Physiology/Medicine Nobel award.
Messenger RNA (mRNA) – a recap on basic biology
Firstly, we should remind ourselves of the structure and key biological role of messenger ribonucleic acid (mRNA). RNA, which comprises nucleosides of 5-carbon sugar ribose linked as phosphodiesters between the 3’- and 5’-hydroxyl groups (the ’ indicating the atoms on the sugar, as non-primed numbers are used to identify the atoms of the bases) is inherently less stable than its much better-known ‘big brother’, DNA. One of the reasons for this is that RNA is much more susceptible to cleavage than DNA. For example, with general base catalysis, RNA can ‘self-cleave’, a process in which the 2-hydroxyl group of the ribose (of course absent in DNA), attacks the neighbouring phosphodiester (Fig. 1). Indeed nature takes advantage of this process in producing molecules called self-cleaving ribozymes. Furthermore, enzymes which cleave RNA – called ribonucleases – are ubiquitous and doggedly stable, and thus handling RNA is generally considered much more difficult than handling DNA.
RNA also differs from DNA in that the 4 different nucleosides used to build it up, i.e. the ‘four letters’, are not exactly the same as in DNA (Fig. 1). In RNA the nucleoside uridine (U) is used instead of the nucleoside thymidine (T). The difference in structure is small, not affecting the H-bonding pattern with the complimentary partner, but quite significant. As we shall see, this year’s Nobel prize is firmly centred on the base uridine (U), and modifications thereof. Thus, the ‘four letters’ in RNA are A, C, G, and U, and U forms a Watson-Crick H-bonded complimentary pair with A, as discussed later.

mRNA is produced from DNA by the process known as transcription; the assembly of the mRNA chains is catalysed by enzymes called RNA polymerases and this happens in the cell’s nucleus, where the DNA is kept. The mRNA then serves as a template encoding the sequence of amino acids that is to be assembled into the protein that is being built, in the process that is known as translation. This is achieved by the cell’s ribosomes, and occurs outside the nucleus in the cell’s much more accessible cytoplasm. Each three letter ‘codon’ either corresponds to one of the 20 proteinogenic amino acids, or as a start or stop signal. Of course this well-known method of conversion of genetic information into protein products is often called the ‘central dogma’ of molecular biology (Fig. 2)

While popular opinion is that all life is based on DNA, in fact many viruses are RNA-viruses; i.e. the genetic material that they contain comprises RNA instead of DNA. SARS-CoV-2 (COVID-19) is probably now the best known example of an RNA virus; it comprises a single strand of RNA which is read in the positive sense in order to produce its viral proteins, and thus is termed a +ssRNA virus. Thus when it replicates, it first converts its RNA into viral proteins, and then it copies its own RNA to replicate itself.
A very simplified account of some basic immunology by a chemist
The human immune system, one of the supreme achievements of biological evolution, has been developing over millions of years to protect us from an enormously diverse and numerous range of pathogens. The immune system is incredibly complicated, and can be very hard to get your head around. However there are a few introductory texts that tackle this difficult task very well.4 It can be broadly split into two parts; firstly the adaptive immune system which is a dynamic system that learns to recognise and remember the structures of invaders. The other part is called the innate immune system, which has slowly developed over a long period of time to recognise general classes of ‘nasties’ and the molecules that are typically associated with them. Vaccines work by stimulating the adaptive immune system, so we will consider how these work in quite general terms next.
Vaccination – tricking the adaptive immune system into being able to defend against future attacks
The basic principle of vaccination is to stimulate an immune response by exposing the person to something that actually poses them no risk, i.e. either a non-infectious version of a virus, or something that looks very much like a virus, but isn’t. The simple idea is that the immune system is turned on to recognise, fight, and destroy this pathogen when it comes across the real thing later on. One really wonders why anti-vaxxers can’t grasp this simple concept?
As stated above, the part of the immune system that does this is called the adaptive immune system. Two of the main types of immune cells that are involved in the adaptive response to infection are T cells and B cells, both of which are types of white blood cell (lymphocyte). B cells produce antibodies, which stick to the outside of invaders, and target them for destruction. T cells, of which there are lots of different sub-types, play several different roles, including activating B cells to produce antibodies, but also in killing attackers themselves.
One of the ways that T cells and B cells can be ‘turned on’ to recognise and help destroy pathogens is by the presentation of short peptides that are derived from the invader on the surface of another type of white blood cell, which are appropriately called antigen presenting cells (APCs). These short peptides are produced by chopping up full-length proteins from the pathogen, for example a viral coat protein, and are normally 8-10 amino acids long. They are displayed by something called MHC class molecules (major histocompatibility complex, types 1 and 2) and are ‘presented’ on the surface of antigen presenting cells for recognition by T cells. Whilst the whole process is complex, it can be simplistically summarised by saying that if APCs present peptides derived from the external proteins of a virus, for example the spike protein of SARS-CoV2, then both T and B cell responses will be mounted to attack and destroy anything that has this viral coat protein.
However it doesn’t stop there. One of the most amazing features of the adaptive immune system is that it ‘remembers’ invaders that it has fought against before, by using ‘memory’ T cells and B cells, so that if it comes across the same pathogen again it can be switched on very quickly to destroy the ‘known’ invader. This memory response may be so effective that actual infection never occurs again, and the person never develops any symptoms.
So, in order for the adaptive immune system to recognise and remember a virus, all it needs to be exposed to are viral proteins. Of course the viral proteins are themselves made from the virus’s own genetic material via the processes of transcription (DNA viruses only) and translation. The obvious advantage of using an mRNA based vaccine over a DNA vaccine is that the transcription step is avoided. In real terms this means that, unlike a DNA vaccine, an mRNA vaccine only needs to access the cytoplasmic ribosomal translation machinery, rather than the cell nucleus. Additionally it does not risk becoming erroneously incorporated into the human’s genome. So, provided the mRNA is translated into protein by the cytoplasmic ribosomes, the viral proteins will be made, chopped up, and displayed on the surface of antigen presenting cells. In this way both memory B and T cell responses will be produced, giving the vaccinated person protection against future infection. So, why therefore have mRNA vaccines not be around for a long time ? The answer lies in how the other part of the immune system, the innate immune system, reacts when it comes across ‘invader’ RNA.
Innate immune response and toll-like receptors (TLRs)
The innate immune system has been evolving over millions of years to protect us from a large range of pathogens. Given that RNA viruses have been around for a very long time, is it hardly surprising that the innate immune system recognises pieces of extraneous RNA as ‘foreign’, and initiates an immune and inflammatory response to destroy them as soon as they are detected. The curiously named ‘toll-like receptors’ (TLRs) are parts of the innate immune system that recognise ‘signature’ molecules of similar structure which are commonly made by a range of different pathogens. Thus they are themselves recognition molecules (in fact large proteins) that generally decorate cell surfaces looking for signs that a type of pathogen may present as a first line of defence.
The German word ‘toll’ can be translated in several ways, but generally means something like ‘fantastic / amazing / crazy’. The prototypic ‘toll receptor’ was discovered in the fruit fly in 1985 by the Nobel Laureates Eric Wieschaus and Christiane Nüsslein-Volhard, and it was the latter who uttered the phrase ‘das ist ja toll’ (that’s so amazing) which led to its naming. They were jointly awarded the Nobel Prize in Physiology/Medicine in 1995 for this discovery. The first human equivalent of this fruit fly receptor was discovered in 1994, and given the name ‘toll-like receptor’ (TLR); the addition of the ‘-like’ therefore simply means that these receptors are human receptors, the function of which mirrors original fruit fly discovery. During the 1990s and 2000s more and more TLRs were discovered, and up to now 10 distinct TLRs have been identified in humans. Indeed these discoveries have been so important in developing our understanding of the immune system that the 2011 Nobel Prize in Physiology/Medicine was also awarded in part for the work on TLRs, on this occasion to Bruce Beutler and Jules Hoffmann.
The point of this distraction is to highlight the fundamental problem with trying to use mRNA as a vaccine in order to develop an immune response against protein. The immune system recognises the foreign RNA (via TLRs 3, 7 and 8) and mounts an immune response against the RNA itself. This means that translation of the mRNA into the actual protein needed for the desired adaptive immune response is very significantly curtailed (if any happens at all), and the vaccination results in an unwanted immune and inflammatory response again the mRNA. So, up until the early 2000s mRNA was commonly regarded as a non-starter for vaccine research, and almost all research effort globally centred on the use of DNA based vaccines, or attenuated viruses. This finally brings us back to the 2023 Nobel prize, which was awarded for finding a way to sneak mRNA past the innate immune system altogether. Let’s now meet the two prize winners.
The 2023 Prize Winners
Drew Weissman – Roberts Family Professor in Vaccine Research and Director of the Institute for RNA Innovations at the University of Pennsylvania

Drew Weissman is an American clinician and immunologist, who was born in Lexington which is just outside Boston in Massachusetts. After receiving his BA and MA degrees in biochemistry and enzymology from Brandeis University, he moved to Boston University where he got a PhD in immunology, in addition to his MD. After a medical residency at the Beth Israel Deaconess Medical Centre in Boston he moved to a Fellowship at the National Institute of Allergy and Infectious Diseases (NIAID), under the guidance of none other than Anthony Fauci who was the Director of NIAID at the time. Fauci was of course later to head the White House task force set up by the Trump administration to deal with the SARS-CoV-2 pandemic.
Weissman’s main research interests were initially centred around HIV and the immune response to infection, and it was in 1997 that he moved to the University of Pennsylvania (UPenn) to establish a lab focussed on studies of RNA and innate immunity. The story goes that he then met his future collaborator Katalin Karikó as they were both trying to use the same photocopier at UPenn, and they immediately clicked, particularly when discussing the difficulty of obtaining funding for RNA research. Weissman was later to say “at the time we couldn't get people to notice RNA as something interesting. Pretty much everybody gave up on it."
Katalin Karikó, Senior vice president at BioNTech Pharmaceuticals and Professor at Szeged University

Professor Katalin Karikó
Katalin Karikó’s journey to the UPenn photocopier was rather more protracted than Drew Weissman’s. She was born into an impoverished family in post-war communist Hungary. Hers is an astonishing and inspirational tale of eventual triumph over adversity that will probably find its way onto cinematic screens over the coming years. There are numerous interviews available to read online5 which provide much more detail that I can give here, and her recently published autobiography6 will no doubt be a fascinating read.
By way of a brief summary, despite being brought up in a house with no running water, no television, nor even a fridge, she excelled at school, and went on to obtain a BSc in biology and then a PhD in biochemistry from the University of Szeged. The research institute in Hungary that she worked in afterwards lost its funding, so she then searched for work abroad and eventually left Hungary to move to the USA with her husband, her two year old daughter, and GBP 900 (purchased on the black market following the sale of their car) which was stuffed inside her daughter’s teddy bear.7
After a postdoc at Temple University she was offered a position at Johns Hopkins, but her postdoc supervisor at Temple objected so strongly to this move that not only did he bad mouth her, but he even reported her to the USA immigration authorities calming that she was ‘illegally in the USA’.8 It took so long for her to successfully appeal the extradition order that was then issued against her that the job offer at Johns Hopkins was subsequently withdrawn.
After a one-year position at the Uniformed Services University of the Health Sciences in Bethesda, Maryland she was eventually hired by the University of Pennsylvania (UPenn), and in 1990 she wrote her first grant application proposing to use mRNA for gene therapy. This, and subsequently more than 20 other grant applications on RNA, were all rejected. In 1995 in a colossal misjudgement, and a move that was later to prove acutely embarrassing, UPenn decided to demote her because of her ‘lack of success in obtaining research grants’. She was thus moved to a ‘non-faculty position, more like a postdoc with no prospect of advancing’.
However, two years later she was to meet Drew Weissman at the faculty photocopier, and immediately they realised that they shared an interest in RNA, and in particular in using synthetic mRNA to potentially develop vaccines. A logical step was to combine their expertise in immunology and molecular biology, and thus began a world-changing and Nobel-winning collaboration. Initially Weissman used one of his NIH grants to support their collaborative mRNA work, even though this grant had no direct link to RNA. Bending the rules a little perhaps, but the work was to prove so vitally important that who would argue with that decision now?
The Nobel-winning breakthrough
As outlined above, the key problem that Karikó and Weissman faced was that in vivo use of mRNA led to severe inflammatory reactions. It was not until 2005 that they were to make the breakthrough for which the Nobel prize was ultimately awarded, and which would later enable such spectacularly successful application of mRNA technology in the COVID-19 vaccines.

To understand that breakthrough we need to look in more detail at one of the nucleosides used to make up RNA, namely uridine, U, and an isomer of that nucleoside called pseudouridine (Ψ, Fig. 3A). Pseudouridine is a C-glycoside; an isomer of uridine in which a carbon atom of the base (uracil) is linked to the anomeric centre of ribose rather than the N1 nitrogen. This is actually a naturally occurring human RNA modification that was first discovered more than 70 years ago,9 and has now been found in many different sorts of RNA (tRNA, rRNA, mRNA, snRNA, etc). It is the most common ‘modified nucleoside’ found in RNA, and there is typically 10 times as much Ψ in human ribosomal RNA as there is in ribosomal bacterial RNA. Moreover, bacterial mRNA contains no Ψ, nor any other modified nucleosides. As shown in Fig 3C, Ψ forms exactly the same Watson-Crick hydrogen bonding base pairing with adenine (A) as uridine (U) does (Fig. 3B). This means that the process of translation of RNA into protein is not affected by replacement of U by Ψ. Interestingly it has been found that incorporation of Ψ into RNA reduces flexibility around the modification,10 favouring stacked and helical conformations, and also provides some protection against cleavage by RNases.
As mentioned previously, avoidance of an innate immune response against the mRNA itself by the toll-like (and other)11 receptors is of critical importance for the efficacy of the vaccines based on mRNA. For many years it was not understood why human RNA is not recognised by TLRs and so is not immunogenic, unlike bacterial and viral RNA. In their breakthrough Nobel-winning 2005 paper, which was published in the journal Immunity after reportedly being rejected by the journals Nature and Science,12 Karikó and Weissman demonstrated that the selective recognition of bacterial RNA by the TLRs was due to the fact that these RNAs contained far fewer ‘structurally modified’ nucleosides. Thus they discovered that marking RNA as ‘self’ by nucleoside modification was the method by which mammalian systems distinguished their own RNA from that of invaders.
Amongst other examples (e.g. methylation of A, C, or U) they showed that incorporating Ψ into mRNA essentially made it undetectable by the innate immune system. Furthermore, they were later to find that the use of Ψ instead of U had other positive and unforeseen consequences. For example, mRNA containing Ψ was translated into protein more efficiently than unmodified mRNA.13 It is thought that this is in part due to reduced binding of the modified mRNA to an RNA dependent protein kinase, and in part due to the increased biological stability14 of mRNA comprising Ψ, allowing greater time for translation.
In later work it was found that replacement of U by N1-methylated pseudouridine (N1MeΨ, Fig. 3A) was even more effective15 in evading the toll-like receptors and also increased translation efficiency further. Furthermore, replacement of the N1-hydrogen atom with a methyl group also removed the possibility of this acting as a H-bond donor (Fig. 3C), and possible ‘wobble-base’ pairing with U, G, or C in the context of a duplex.16 All of the current COVID-19 vaccines therefore contain N1MeΨ17 at all positions where a U would have been expected based on the COVID-19 spike protein sequence.
The COVID-19 vaccines: what do they contain and how are they made?
Overcoming the toll-like receptor problem was not the only obstacle to developing effective mRNA vaccines. Other key advances were also essential so that the vaccine was able to reach its target by injection into muscle. Probably the most important of these was the development of an ingenious lipid nanoparticle formulation,18 which protected the mRNA from degradation by RNases, ensured cellular penetration, and also acted as an adjuvant (something that increases the immune response to a vaccine). Cellular penetration of naked mRNA is negligible, and putting mRNA inside lipid nanoparticles was shown to increase uptake and expression more than 1000-fold compared to naked mRNA in animal models.19 Structural studies have shown that each lipid nanoparticle contains a low number of copies of the mRNA, typically 1-10.
The modified mRNA that makes up the COVID-19 vaccines is generated from a DNA template by in vitro transcription in the presence of N1-methylpseudouridine-5′-triphosphate (N1MeΨTP), instead of the normal uridine-5′-triphosphate (UTP). Synthetic RNA made in this way is often called in vitro transcribed mRNA; IVT mRNA for short. Because this manufacturing process does not involve cell lines, it is much faster and cheaper than alternative methods of vaccine production, for example using a killed or attenuated live virus. In addition to coding the sequence of the SARS-CoV2 spike protein, the vaccine mRNA also contains a few extra elements, such as a 5’-cap and a 3’-poly-A (adenosine) tail which mimic natural mRNA, and also some sequence elements that further increase RNA stability and translation efficiency in human antigen presenting cells. Typically, ~100 copies of mRNA are made per molecule of the DNA template, so this in vitro translation approach is amenable to large scale production; practically this is achieved using an RNA polymerase for transcription, which transcribes the template DNA into mRNA only downstream of a specific promoter site (for example T7). Finally, the mRNA encoding the spike protein is formulated with the lipids, polyethylene glycol (PEG), sucrose, and some salts, to make the mRNA–lipid nanoparticle drug that is administered by intramuscular injection.
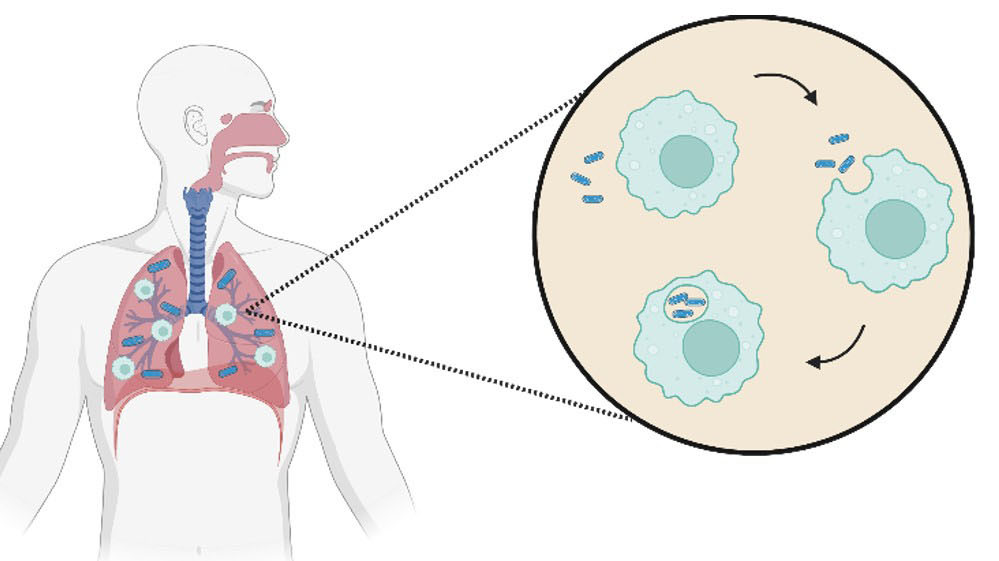
It is currently thought that muscle cells are not very effective at producing the spike protein. Rather what happens is that the nanoparticles make their way to proximal lymph nodes where they are internalised by dendritic cells, which are the most important type of antigen presenting cells (APCs). Thus, following internalisation, the mRNA package is released from the lipid nanoparticle, and is then translated by the ribosomes to form the virus spike protein (Fig 4). This spike protein is fully folded and properly post-translationally modified, for example by glycosylation. These dendritic cells then display the cut-up spike protein peptides on their surfaces and activate and induce the B and T cells responses, leading in turn to antibody and cytokine production, and most important of all memory T and B cell production.

What does the future hold ?
The beauty of the mRNA system is that it is almost infinitely adaptable.20 Simply changing the sequence of nucelosides in the mRNA will change the sequence of amino acids in the immunogenic protein that will be produced. With respect to the rapidly changing face of SARS-CoV2 this means that as the virus changes its spike protein structure, so the vaccine can be changed. Indeed most of us will now have had second or third generation bivalent versions of the Pfizer/BioNTech or Moderna vaccines which also contain mRNA encoding Omicron strains of the virus, such as BA.4 and BA.5.
So, the sky is the limit you may say? Could we perhaps have mRNA vaccines against many other viruses in the near future? Well, mRNA vaccines against viruses such as HIV, Zika and rabies are already in development. And that’s not all – could we produce an immune response against other cells displaying particular proteins, or protein fragments, on their surface? For example, how about cancer vaccines? Why don't we turn the immune system on to kill our own cells that are over-expressing proteins (antigens) that are associated with cancer, for example growth associated factors or proteins that are unique to tumour cells due to mutations? Here indeed would be Ehrlich’s ‘magic bullet’ – a way to specifically and selectively kill only one rogue cell type amongst millions of healthy others. Although only one cancer vaccine has so far been approved by the FDA, there are numerous clinical trials now being performed using mRNA vaccines21 with just this goal in mind, including recent entries into Phase 3.22
In fact Karikó and Weissman’s initial work on mRNA actually didn't mention vaccines at all; rather that early work was aimed at using mRNA for ‘gene therapy’, something that had first been reported using mRNA as far back as 1990, but with no follow up.23 To understand this application we need to remember that genes encode proteins. The idea here then is to enable people who have a faulty or missing gene to be able to produce the protein that they lack by infecting their cells with mRNA, which will then in turn be translated into that missing protein. Gene therapy is indeed quite an old idea, though current approaches usually involve infecting the sufferer with a live virus, such as lentivirus or adenovirus, which encodes the faulty or missing gene. At the time of writing there are currently eleven such gene therapy treatments that have already been approved by the FDA and many more are in trials. However, mRNA-based gene therapy approaches may have some advantages over these DNA-based approaches, including the ease and scalability of production, and the limited lifetime of the mRNA which in turn provides the ability to rapidly regulate the amount of protein produced.
As of yet none of these other applications for mRNA are yet in place; the COVID-19 vaccine program was of course accelerated quite remarkably due to the critical global situation. However, we can certainly expect to see many more ground-breaking applications of mRNA technology in the coming years; all of it relying on the use of pseudouridine to avoid detection by the innate immune system.
Afterthoughts
Given the devastating effects of the COVID-19 pandemic, and the massive beneficial impact that the mRNA vaccine technology has already had, it is no surprise that Karikó and Weissman were awarded a Nobel Prize. Readers of my previous two articles will understand that my surprise was perhaps that they should have won one sooner. Moreover the ‘rags to riches’ story of Katalin Karikó is quite remarkable: her triumph over adversity and repeated rejection, her amazing determination and self-belief. It is an inspirational tale to be told to all future scientists.
Finally, I should add that the 2-year-old little girl, inside whose teddy bear Katalin Karikó hid their black-market £s as they left Hungary, is now called Susan (Zsuzsanna) Francia. She is a two-time Olympic Gold medallist rower, winning in the women’s eight in both 2008 and 2012. So, perhaps there is a gene for determination and success? If so, then maybe someone could give me a dose of an mRNA gene therapy encoding it?
References and notes
- https://www.youtube.com/watch?v=0JP-yDsLc3k
- Fairbanks, A.J. Chem. NZ 2022, 86, 10-14.
- Fairbanks, A.J. Chem. NZ 2023, 87, 18-27.
- For a highly recommended introductory text see: ‘How the Immune System Works’, Lauren Sompayrac, 7th Edition, Wiley Blackwell, 2022, ISBN 9781119890683.
- For links to many online interviews and further details see: https://en.wikipedia.org/wiki/Katalin_Karikó.
- ‘Breaking Through; My Life in Science’, Katalin Karikó, Penguin Random House, 2023, ISBN 9780593443163.
- https://www.washingtonpost.com/health/2021/10/01/katalin-kariko-covid-vaccines/.
- ‘A Shot to Save the World: The Inside Story of the Life-or-Death Race for a COVID-19 Vaccine’, Gregory Zuckerman; Penguin Random House, 2021, ISBN 9780593420393.
- Cohn, W.E.; Volkin, E. Nature 1951, 483-484.
- Westhof, E. Biochem. Biophys. Res. Commun. 2019, 520, 702–704.
- The immune system is incredibly complicated, and unsurprisingly does have other receptors to mount an innate immune response against single and double stranded RNA. For further details see: Tatematsu, M.; Funami, K.; Seya, T.; Matsumoto, M. J. Innate Immun. 2018, 10, 398–406.
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Immunity 2005, 23, 165–175.
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Mol. Ther. 2008, 16, 1833–1840.
- Thought to be due to a combination of increased resistance to phosphodiesterases, and increased stability of RNA secondary structures; Y promotes base stacking. See Ref 13.
- Andries, O.; Cafferty, S.M.; Smedt, S.C.D.; Weiss, R.; Sanders, N.N.; Kitada, T. J. Control. Release 2015, 217, 337–344.
- Kierzek, E.; Malgowska, M.; Lisowiec, J.; Turner, D.H.; Gdaniec, Z.; Kierzek, R. Nucleic Acids Res. 2014, 42, 3492–3501.
- For a more in-depth discussion of the importance of pseudouridine in mRNA COVID-19 vaccine development see: Morais, H. Adachi, Y.-T. Yu, Front. Cell Dev. Biol. 2021, 9, 789427.
- Buschmann, M.D.; Carrasco, M.J.; Alishetty, S.; Paige, M.; Alameh, M.G.; Weissman, D. Vaccines 2021, 9, 65.
- Oberli, M.A.; Reichmuth, A.M.; Dorkin, J.R.; Mitchell, M.J.; Fenton, O.S.; Jaklenec, A.; Anderson, D.G.; Langer, R.; Blankschtein, D. Nano Lett. 2017, 17, 1326–1335.
- For a review written before the advent of COVID-19 which considers future applications of mRNA vaccines see: Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. Nat. Rev. Drug Discov. 2018, 17, 261–279.
- Wang, B.; Pei, J.; Xu, S.; Liu, J.; Yu, J. Front. Immunol. 2023, 14, 1246682.
- Carvalho, T. Nat. Med. 2023, 29, 2379–2380.
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani A. et al. Science 1990, 247, 1465-1468.